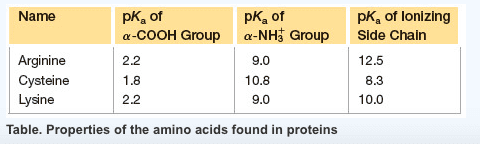
1. What is the optimum pH to separate a mixture of lysine, arginine, and cysteine using electrophoresis? (See the table in the image.)
A. At pH about 7.0
B. At pH about 5.1
C. At pH about 9.5
D. At pH about 10.8
2. Choose the structure of lysine in the protonation state that would predominate at the pH you have chosen.
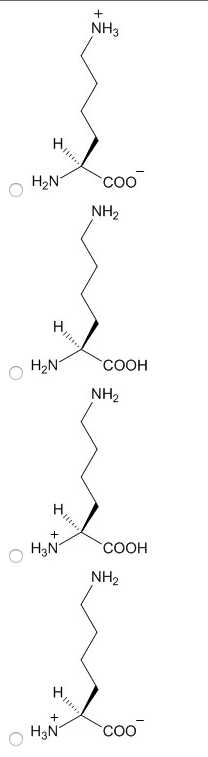
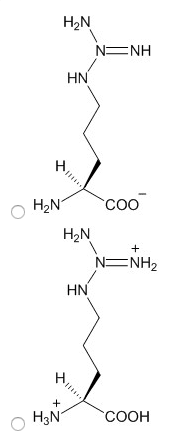
3. Choose the structure of arginine in the protonation state that would predominate at the pH you have chosen.
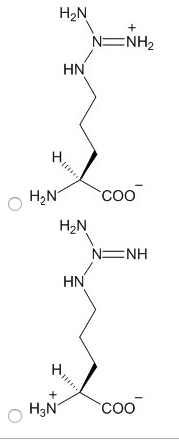
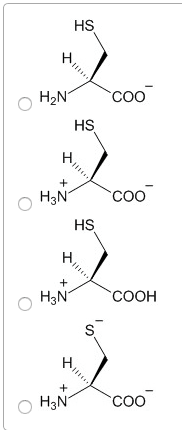
4. Choose the structure of cysteine in the protonation state that would predominate at the pH you have chosen.
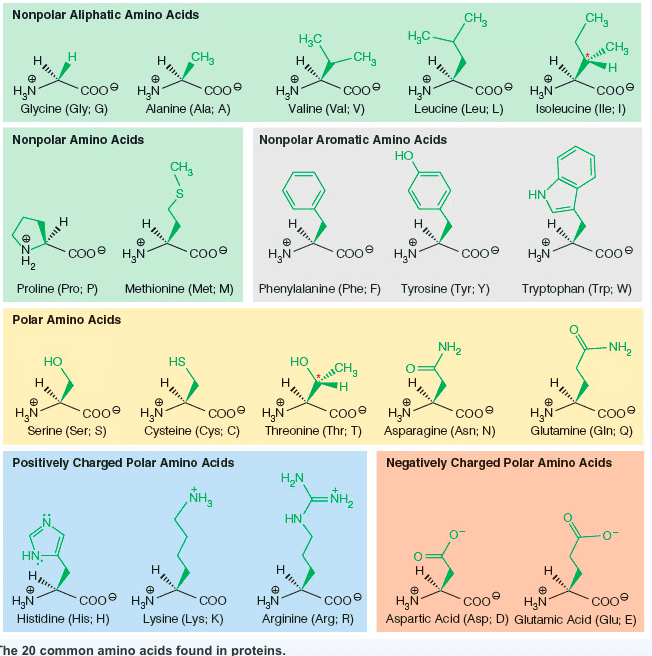
Nonpolar Aliphatic Amino Acids CH Hac CH H. H H. CH2 CH COoHaN Glycine (Gly: G) Alanine (Ala; A) Valine (Val; V) Leucine (Leu; L) Isoleucine (lle; I) Nonpolar Amino Acids Nonpolar Aromatic Amino Acids cooe H&N H,N COO Î Proline (Pro; P) Methionine (Met; M) Phenylalanine (Phe; F) Tyrosine (Tyr; Y) Tryptophan (Trp: W) Polar Amino Acids cooe HN cooe HN H2N Glutamine (Gln; Q) H,N cooe H.N Serine (Ser; S) Cysteine (Cys; C) Threonine (Thr; T) Asparagine (Asn; N) Positively Charged Polar Amino Acids Negatively Charged Polar Amino Acids NH HN COo H3N COO Î a H3N Histidine (His; H) Lysine (Lys; K) Arginine (Arg: R)Aspartic Acid (Asp; D) Glutamic Acid (Glu; E) he 20 common amino acids found in proteins







